目录
背景
- 可变剪切事件的检测需要很好的测序丰度
- 现在有大量的公共RNAseq数据集,是可以提供可变剪切的信息的
- 对于低coverage的样本,其可变剪切事件通常是检测不出来的。有效的检测可变剪切事件,以及不同样本间的差异,没有特别好的方法
模型
- DARTS:Deep-learning Augmented RNAseq analysis of Transcript Splicing
- 模型:
- 包含两个部分:
- DNN:基于序列特征和RBP的表达量特征,学习差异可变剪切的潜在模型。该模型作为大规模数据集的先验知识用于新样本的推断。
- BHT:贝叶斯假设检验模块。这里包含了两个,一个是BHT(flat),是之前他们的预测剪切事件的工具(如rMATS)的升级版,这里主要用来对于大规模的公共数据集(如ENCODE、Roadmap)进行分析,得到其剪切事件的结果,构建一个具有label的数据集用于DNN训练。还有一个模块是BHT(info),把要分析的新的样本作为样本特征,DNN的预测作为先验知识,对新样本的可变剪切进行分析。
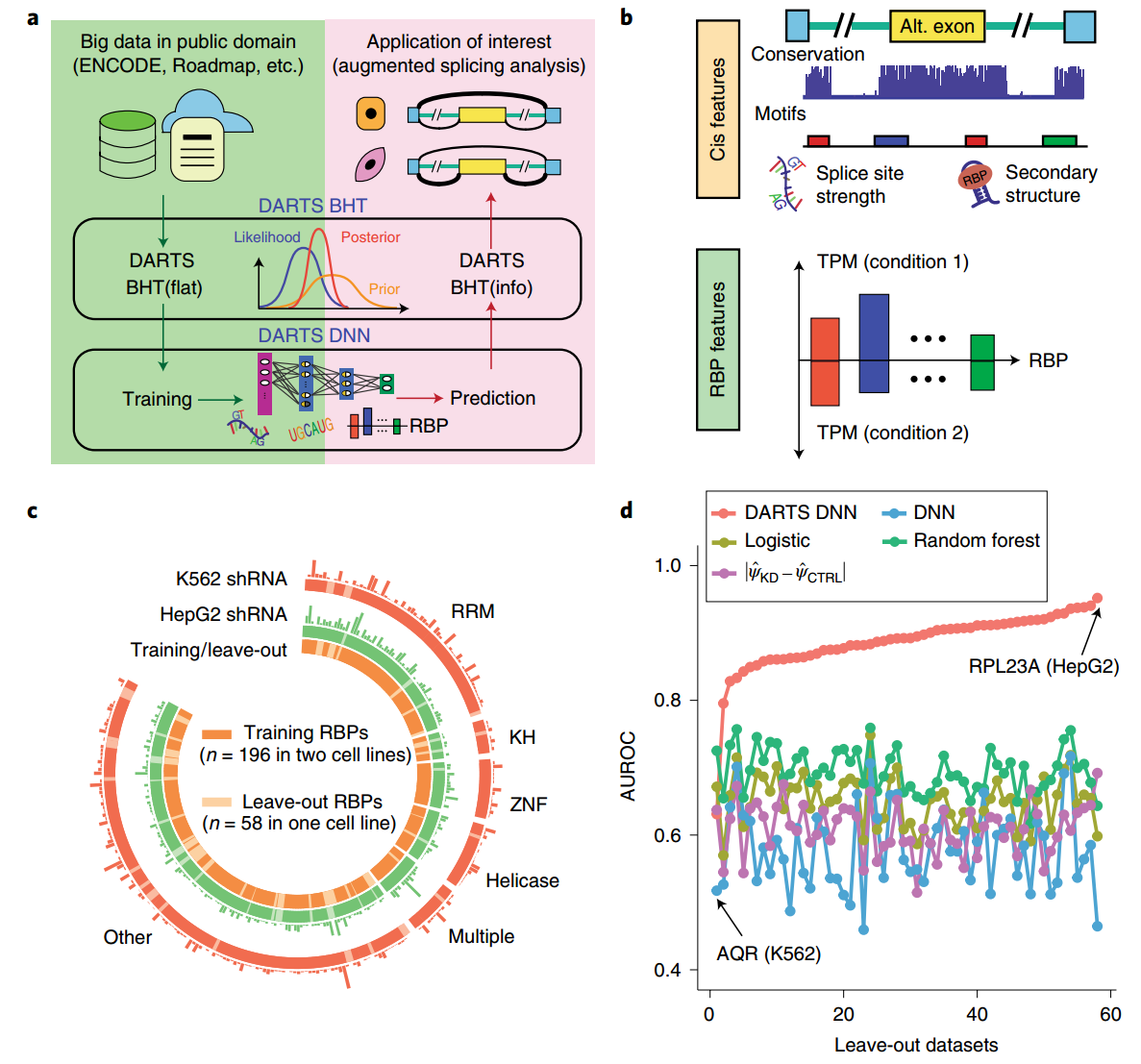
- a)模型的基本框架
- b)在DNN模型中,用到的特征,包括序列特征(cis),和RBP表达量的特征(trans)
- c)DNN训练时,使用的数据集。这里关注的是差异可变剪切,所以用的都是shRNA KD vs control的样本对,两个不同的细胞系(K562+HepG2)
- d)模型效果的比较。图中的DNN是在单个数据集上训练的,DARTS-DNN是随着横轴增大,在对应数目上的数据集上训练的。还测试另外的模型:逻辑回归,随机森林,还有一个直接预测PSI然后取两个条件的差值的方式。综合来看,DARTS-DNN效果是最好的,但是需要在多个数据集上进行训练。
效果评估
应用测试
- 应用1:EMT过程
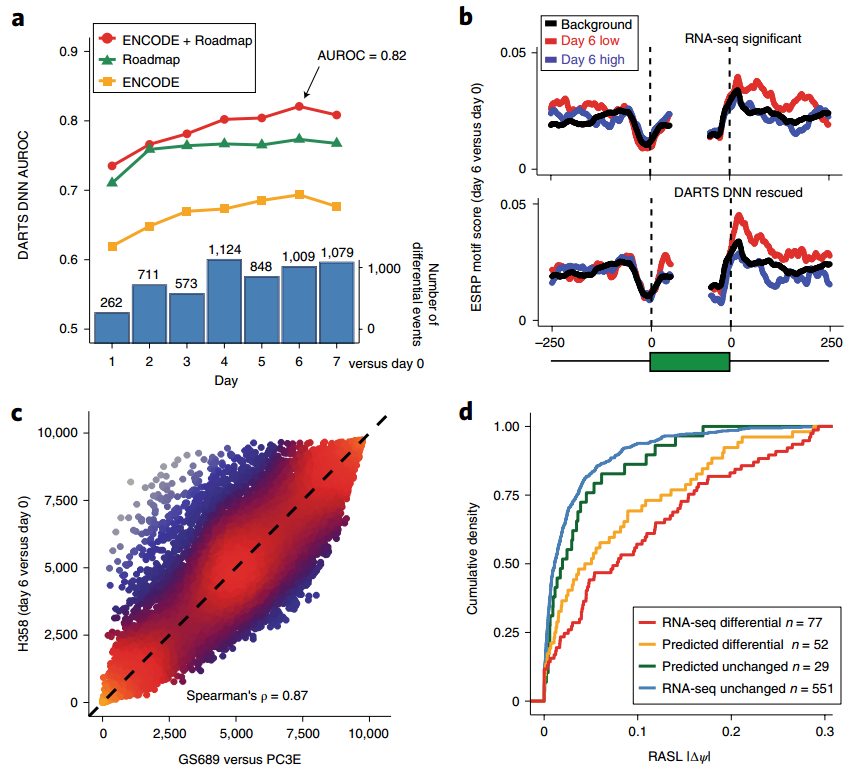
- 7个时间点,都和第0天比较,看差异可变剪切的变化
-
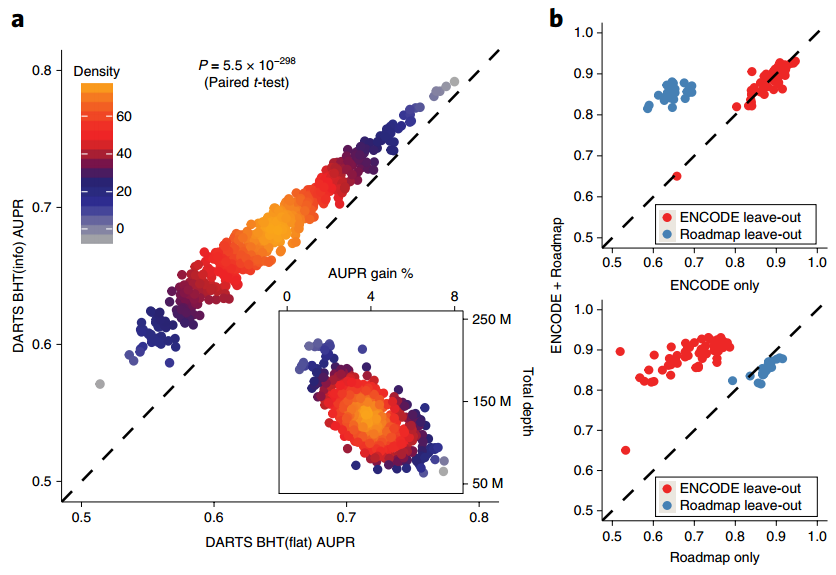
DNN训练来源不同,可以分为不同的模型,可以看到整合的来源最后预测的效果是最好的(图a)
- DARTS-DNN rescue
-
使用两个模型:BHT(flat)、DARTS-DNN,比较第6天和第0天得到差异可变剪切,再看在exon-intron junction区域的motif的富集(图b)
- 使用两个不同的系统去看EMT过程
- 但是分别得到的效果是高度正相关的,说明模型的鲁棒性(图c)
- 应用2:RASL-seq低表达量转录本
- 特异的,检测低表达的转录本的差异可变剪切事件
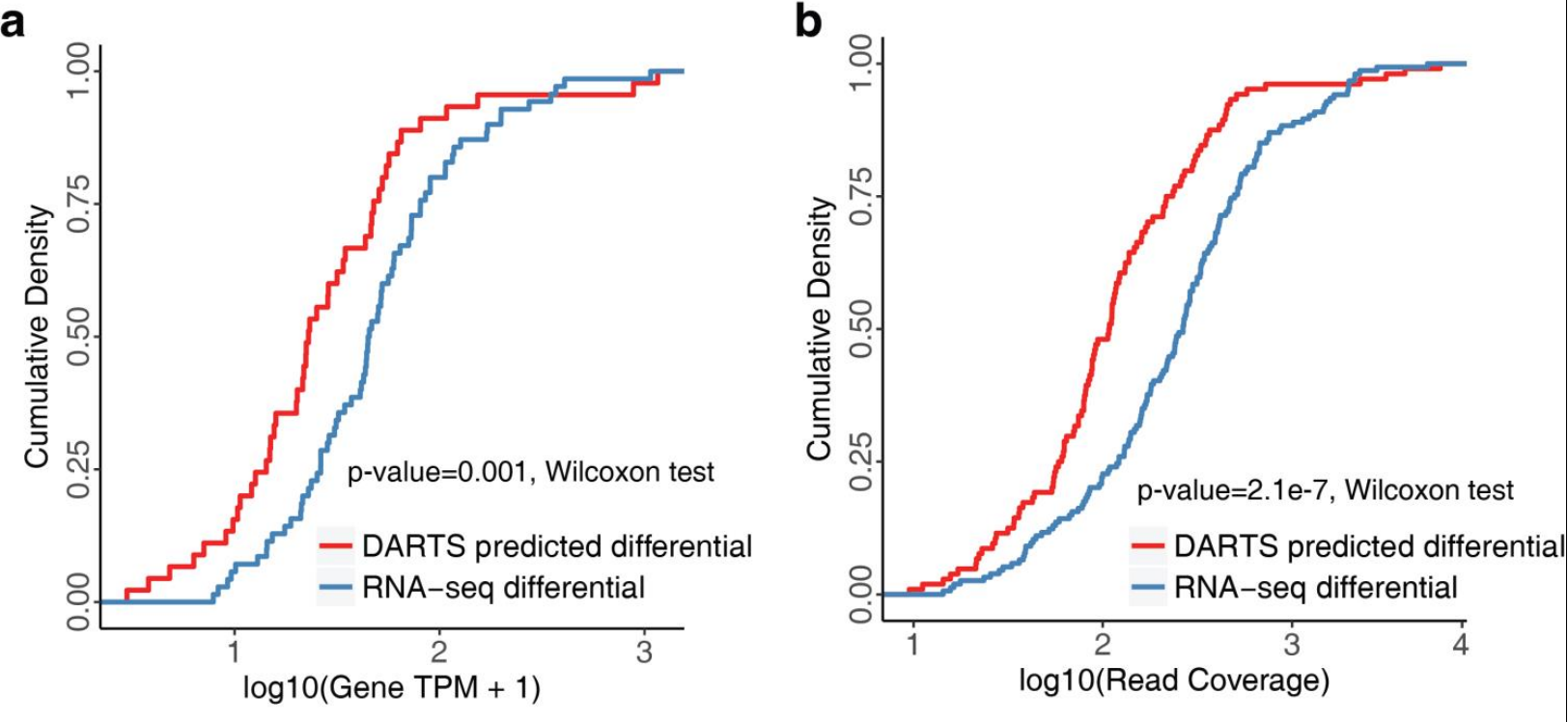
- 对于图d,这么来看。首先用BHT(flat)直接去预测,得到了差异剪切(RNA-seq differential)的和不变的(RNA-seq unchanged),累计曲线显示前者比后者的score大很多,符合预期。接下来用DARTS-DNN辅助预测,能够预测到一些新的,就是图中的预测的差异剪切(predicted differential)和不变的(predicted unchanged),可以看到这两个的趋势跟上面的是一致的,也就是差异的score比不变的大很多。单独看对应类型的(比如RNA-seq differential vs predicted differential),其曲线也是很接近的。在补充材料里面,其实看了这两个类型的转录本的表达量水平,发现新增预测到的其实表达量是更低的(如下图的ab,分别看的是表达量水平和read coverage)。由此说明,对于低表达量的转录本,DARTS也是可以很好的检测到差异可变剪切事件的。
代码
相关的代码已经放在 Xinglab/DARTS 上面了,可以参考。
- Darts_DNN@github
- Darts_BHT@github
- Darts_BHT&DNN@readoc
- Trained models and related files@sourceforge:有很多文件需要从这里下载,包含四个不同的模型:SE(skipped exons),RI(retained introns),A5SS(alternative 5’ splice sites),A3SS(alternative 3’ splice sites)。根据自己要预测的事件类型,选取对应的已训练的模型参数和文件。不同类型事件的序列特征数目不同,RBP表达特征数目是相同的。
DNN predict commad:
➜ bin git:(master) ✗ python Darts_DNN predict -h
usage: Darts_DNN predict [-h] -i INPUT -o OUTPUT [-t {SE,A5SS,A3SS,RI}]
[-e EXPR [EXPR ...]] [-m MODEL]
optional arguments:
-h, --help show this help message and exit
-i INPUT Input feature file (*.h5) or Darts_BHT output (*.txt)
-o OUTPUT Output filename
-t {SE,A5SS,A3SS,RI} Optional, default SE: specify the alternative splicing
event type. SE: skipped exons, A3SS: alternative 3
splice sites, A5SS: alternative 5 splice sites, RI:
retained introns
-e EXPR [EXPR ...] Optional, required if input is Darts_BHT output;
Folder path for Kallisto expression files; e.g '-e
Ctrl_rep1,Ctrl_rep2 KD_rep1,KD_rep2'
-m MODEL Optional, default using current version model in user
home directory: Filepath for a specific model
parameter file
DNN prediction:
# 需要先下载model文件:master.dl.sourceforge.net/project/rna-darts/resources/DNN/v0.1.0/trainedParam/A5SS-trainedParam-EncodeRoadmap.h5
Darts_DNN predict -i darts_bht.flat.txt -e RBP_tpm.txt -o pred.txt -t A5SS
darts_bht.flat.txt:
➜ test_data git:(master) ✗ head darts_bht.flat.txt
ID I1 S1 I2 S2 inc_len skp_len mu.mle delta.mle post_pr
chr1:-:10002681:10002840:10002738:10002840:9996576:9996685 581 0 462 0 155 99 1 0 0
chr1:-:100176361:100176505:100176389:100176505:100174753:100174815 28 0 49 2 126 99 1 -0.0493827160493827 0.248
chr1:-:109556441:109556547:109556462:109556547:109553537:109554340 2 37 0 81 119 99 0.0430341230167355 -0.0430341230167355 0.188
chr1:-:11009680:11009871:11009758:11009871:11007699:11008901 11 2 49 4 176 99 0.755725190839695 0.117542135892979 0.329333333333333
chr1:-:11137386:11137500:11137421:11137500:11136898:11137005 80 750 64 738 133 99 0.0735580941766509 -0.0129207126090368 0
chr1:-:113247674:113247790:113247721:113247790:113246265:113246428 159 1862 127 1958 145 99 0.0550902772187827 -0.0126831240261882 0
chr1:-:1139413:1139616:1139434:1139616:1139223:1139348 980 106 24 2 119 99 0.884944451538756 0.0240073464719553 0.128666666666667
chr1:-:115166127:115166264:115166180:115166264:115165607:115165720 17 0 32 1 151 99 1 -0.0454956312142212 0.287333333333333
chr1:-:11736865:11737005:11736904:11737005:11736102:11736197 22 1532 26 1544 137 99 0.0102705812451076 0.00175172587953396 0
RBP_tpm.txt:
➜ test_data git:(master) ✗ head RBP_tpm.txt
thymus adipose
RPS11 2678.83013 2531.887535
ERAL1 14.350975 13.709394
DDX27 18.2573 14.02368
DEK 32.463558 14.520312
PSMA6 102.332592 77.089475
TRIM56 4.519675 6.14762566667
TRIM71 0.082009 0.0153936666667
UPF2 7.150812 5.23628033333
FARS2 6.332831 7.291382
参考
- How Does a New Computational Method Transform Public Big Data Into Knowledge of Transcript Splicing
- New computational tool harnesses big data, deep learning to reveal dark matter of the transcriptome
- 用深度学习预测可变剪切
- 邢毅团队利用深度学习强化RNA可变剪接分析的准确性
If you link this blog, please refer to this page, thanks!
Post link:https://tsinghua-gongjing.github.io/posts/DARTS.html
Previous:
Excel常见用法
Next:
使用迁移学习对scRNA数据降噪
Latest articles
Links
- ZhangLab , RISE database , THU life , THU info
- Data analysis: pandas , numpy , scipy
- ML/DL: sklearn , sklearn(中文) , pytorch
- Visualization: seaborn , matplotlib , gallery
- Github: me