file format
header lines
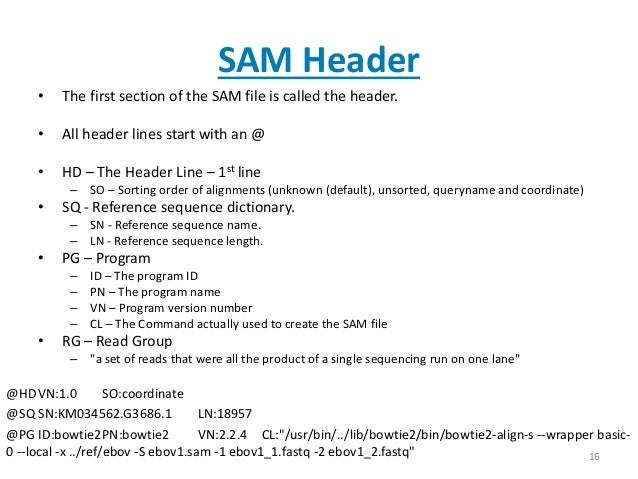
meaning of each column:
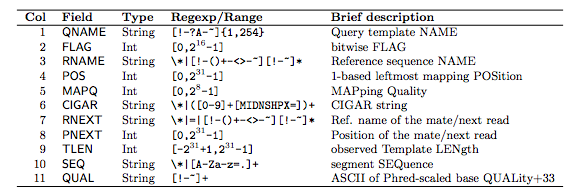
FLAG: how is the read mapped ?
explain-flags from broad institue
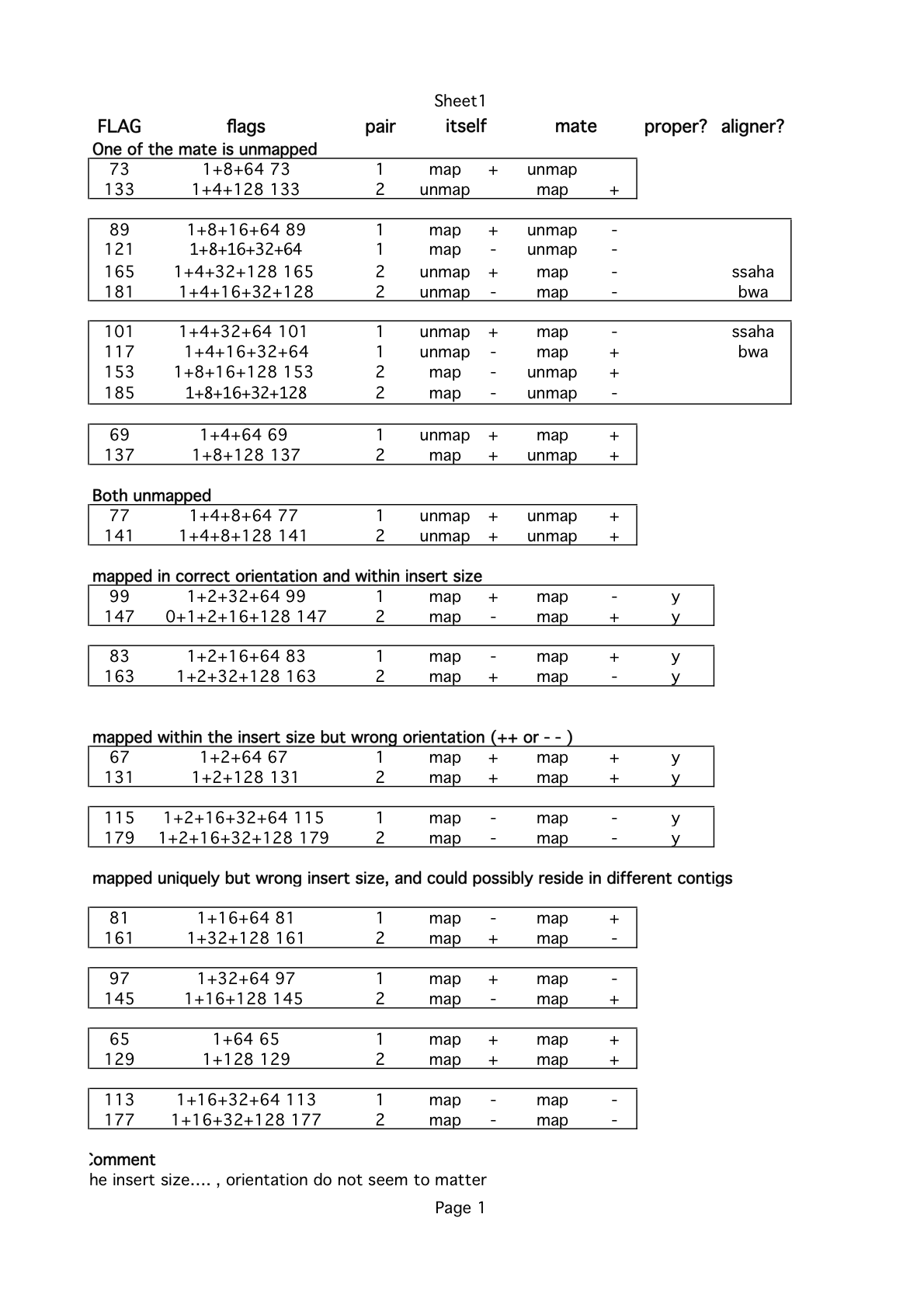
Example operation
Here SAM and BAM filtering oneliners (github) list some of basic usages
Extracting reads for a single chromosome from BAM/SAM source
If no regions or options: print all, If specific one or more regions (space separted): print restricted Note: need sorted and indexed
samtools view -h hur_MO-h6_rep2.sorted.bam NM_001002366 > test.sam
# convert bam directly
samtools view -bS HG00096.chr20.sam > HG00096.chr20.bam
# specific multiple chromosome or regions by space
samtools view -bS *bam chr1 chr2 chr3 > test.bam
Calculate number of mapped reads biostar
# number of entries excluding those marked as read #1 in a pair
[zhangqf7@bnode02 decay]$ samtools view -F 0x40 RIP_h4.bam| cut -f1 | sort | uniq | wc -l
104336
[zhangqf7@bnode02 decay]$ samtools flagstat RIP_h4.bam
208877 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
208877 + 0 mapped (100.00% : N/A)
208877 + 0 paired in sequencing
104541 + 0 read1
104336 + 0 read2
200984 + 0 properly paired (96.22% : N/A)
207883 + 0 with itself and mate mapped
994 + 0 singletons (0.48% : N/A)
71 + 0 with mate mapped to a different chr
71 + 0 with mate mapped to a different chr (mapQ>=5)
# Count mapped reads number from sam file
[zhangqf7@ZIO01 bowtie2_genome]$ samtools flagstat riboseq.sam
1704656 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
1704656 + 0 mapped (100.00% : N/A)
0 + 0 paired in sequencing
0 + 0 read1
0 + 0 read2
0 + 0 properly paired (N/A : N/A)
0 + 0 with itself and mate mapped
0 + 0 singletons (N/A : N/A)
0 + 0 with mate mapped to a different chr
0 + 0 with mate mapped to a different chr (mapQ>=5)
# for single-end data
[zhangqf7@bnode02 decay]$ samtools view -F 0x904 RIP_h4.bam| cut -f1 | sort | uniq | wc -l
104971
# for pair-end data
[zhangqf7@bnode02 decay]$ samtools view -F 0x4 RIP_h4.bam| cut -f1 | sort | uniq | wc -l
104971
Compute coverage of each base
bamCoverage from deeptools [slow]
# old version
bamCoverage --bam RIP_h6.bam --outFileName RIP_h6.bam.bw --binSize 1 --normalizeTo1x 37421801
# 37421801, the total bases in our merge reference
# new version of bamCoverage (DeepTools)
bamCoverage --bam RIP_h6.bam --outFileName RIP_h6.bam.bw --binSize 1 --effectiveGenomeSize 37421801 --normalizeUsing RPGC
# RPGC: RPGC (per bin) = number of reads per bin / scaling factor for 1x average coverage. This scaling factor, in turn, is determined from the sequencing depth: (total number of mapped reads * fragment length) / effective genome size. The scaling factor used is the inverse of the sequencing depth computed for the sample to match the 1x coverage. This option requires –effectiveGenomeSize.
igvtools count command in igvtools [fast]
igvtools count -w 1 RIP_h4.bam RIP_h4.wig /Share/home/zhangqf7/gongjing/zebrafish/data/reference/gtf/refseq_ensembl91_merge.tarns.fa
Add tag using pysam
ref: biostar
r = pysam.AlignedRead()
# add alignment end ad tag 'EN' which can be used for color
r.tags += [('EN', read.reference_end)]
If you link this blog, please refer to this page, thanks!
Post link:https://tsinghua-gongjing.github.io/posts/sam-file.html
Previous:
Python module pandas
Next:
Heatmap plot
Latest articles
Links
- ZhangLab , RISE database , THU life , THU info
- Data analysis: pandas , numpy , scipy
- ML/DL: sklearn , sklearn(中文) , pytorch
- Visualization: seaborn , matplotlib , gallery
- Github: me