使用IGV加载bed文件
command line: igvtools
% ~/Downloads/IGVTools/igvtools help
Program: igvtools. IGV Version 2.3.98 (141)07/25/2017 12:12 AM
Usage: igvtools [command] [options] [input file/dir] [other arguments]
Command: version print the version number
sort sort an alignment file by start position.
index index an alignment file
toTDF convert an input file (cn, gct, wig) to tiled data format (tdf)
count compute coverage density for an alignment file
formatexp center, scale, and log2 normalize an expression file
gui Start the gui
help <command> display this help message, or help on a specific command
See http://www.broadinstitute.org/software/igv/igvtools_commandline for more detailed help
Done
sort and index
在加载之前,先用igvtools建立索引文件,尤其是对于比较大的bed文件,这样能节省加载时所耗用的内存,否则在加载多个track时容易出现加载失败。
在建立inde之前,需要sort bed文件,有两种方式:
sort -k1,1 -k2,2n in.bed > out.sort.bed (bedtools 推荐)
sort -k1,1 -k2,2n -o in.bed in.bed # 直接sort原文件并保存
igvtools sort in.bed out.sort.bed
igvtools index sort.bed
load into IGV
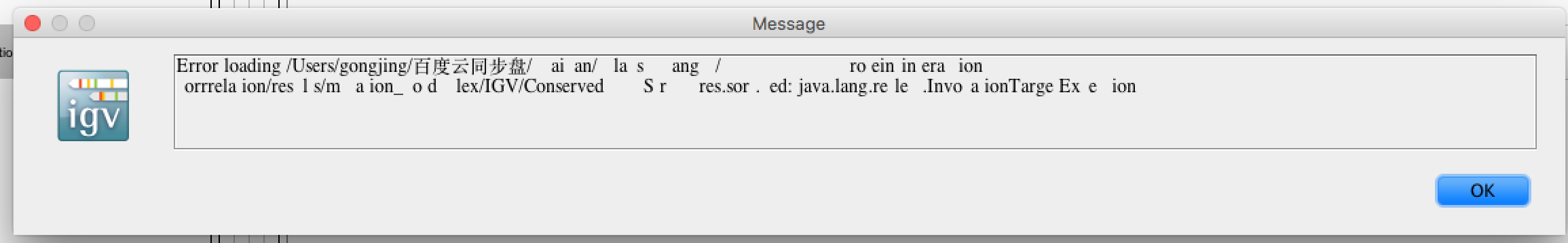
在直接load进去的时候,出现报错:
原因是:所建立的index文件的路径含有中文字符,而这貌似是不支持的,在google的igv-help的群组里面也有类似的问题如下:
load bed file with color
The .bed file should be defined like this, the 5 column should not be ., must specify a value.
track name="ItemRGBDemo" description="Item RGB demonstration" visibility=2 itemRgb="On"
chr1 629885 629939 IGF2BP3 1 + 629885 629939 255,250,200
chr1 629887 629940 IGF2BP1 1 + 629887 629940 170,110,40
chr1 629891 629928 NOP56 1 + 629891 629928 0,0,0
chr1 629891 629931 FBL 1 + 629891 629931 60,180,75
If you link this blog, please refer to this page, thanks!
Post link:https://tsinghua-gongjing.github.io/posts/IGV_load_bed_with_index.html
Previous:
Collected cheatsheet for quick reference
Next:
Joint plot
Latest articles
Links
- ZhangLab , RISE database , THU life , THU info
- Data analysis: pandas , numpy , scipy
- ML/DL: sklearn , sklearn(中文) , pytorch
- Visualization: seaborn , matplotlib , gallery
- Github: me