目录
bed file
Full description can be accessed at UCSC bed, here are example from bedtools introduction :
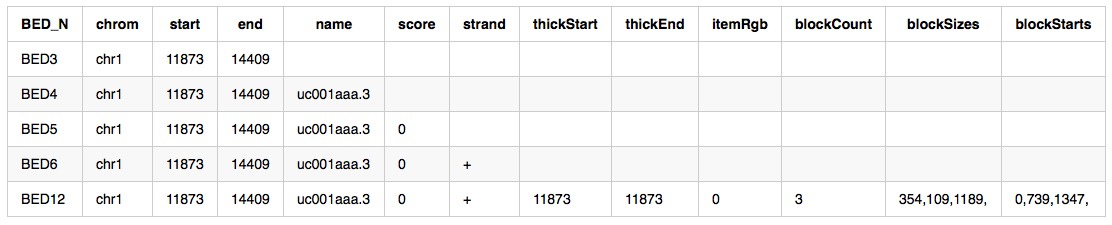
columns: 12 (some are optional correspond to different style)
- chrom - The name of the chromosome on which the genome feature exists.
- start - The 0-based starting position of the feature in the chromosome.
- end - The one-based ending position of the feature in the chromosome.
- name - Defines the name of the BED feature.
- score - The UCSC definition requires that a BED score range from 0 to 1000, inclusive.
- strand - Defines the strand - either ‘+’ or ‘-‘.
- thickStart - The starting position at which the feature is drawn thickly.
- thickEnd - The ending position at which the feature is drawn thickly.
- itemRgb - An RGB value of the form R,G,B (e.g. 255,0,0).
- blockCount - The number of blocks (exons) in the BED line.
- blockSizes - A comma-separated list of the block sizes.
- blockStarts - A comma-separated list of block starts.
wig & bigwig file
- UCSC bigWig Track Format: https://genome.ucsc.edu/goldenpath/help/bigWig.html
- for dense, continuous data
- bigWig:
- indexed binary
- faster display performance
# convert .bw to .wig
bigWigToWig bigWigExample.bw out.wig
# large bedgraph to .bw
bedGraphToBigWig in.bedGraph chrom.sizes myBigWig.bw
Example:
$ head AGN001508.bedGraph AGN001508.wig
==> AGN001508.bedGraph <==
chr1 1720 1752 2.99808
chr1 6751 6760 2.99808
chr1 6891 6916 2.99808
chr1 13926 13969 2.99808
chr1 14504 14537 2.99808
chr1 14545 14555 2.99808
chr1 14555 14584 5.99616
chr1 14584 14586 2.99808
chr1 14586 14588 5.99616
chr1 14588 14596 2.99808
==> AGN001508.wig <==
#bedGraph section chr1:1720-25052994
chr1 1720 1752 2.99808
chr1 6751 6760 2.99808
chr1 6891 6916 2.99808
chr1 13926 13969 2.99808
chr1 14504 14537 2.99808
chr1 14545 14555 2.99808
chr1 14555 14584 5.99616
chr1 14584 14586 2.99808
chr1 14586 14588 5.99616
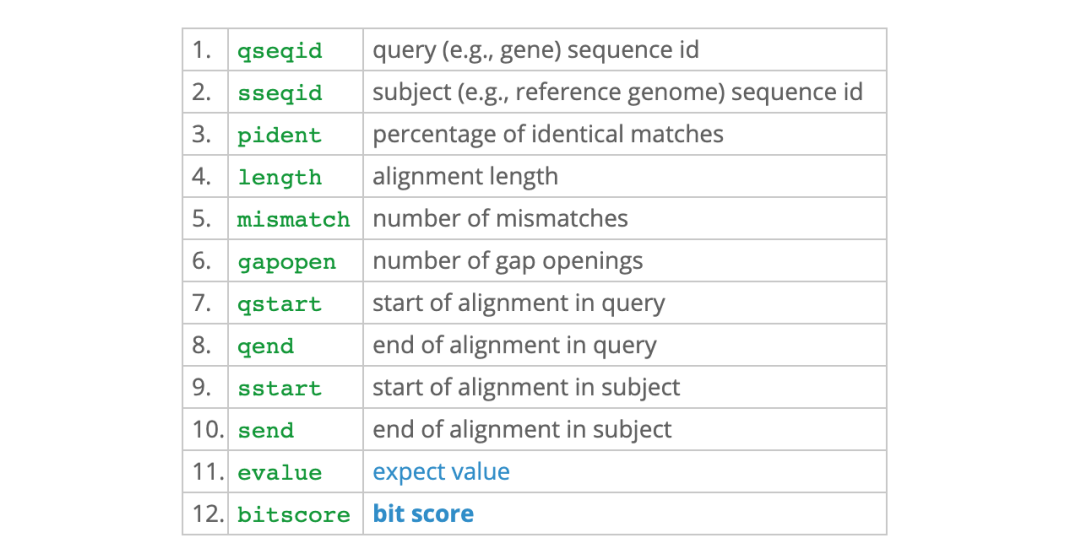
blastn outfput format6
➜ seq_similarity head -3 outputfile_E10
ENST00000380087:1800-1900 ENST00000330735:600-700 100.00 66 0 0 166 35 100 1e-29 122
ENST00000380087:1800-1900 ENST00000330735:700-800 100.00 34 0 0 67100 1 34 6e-12 63.9
ENST00000557530:200-300 ENST00000348956:500-600 100.00 93 0 0 1 938 100 1e-44 172
If you link this blog, please refer to this page, thanks!
Post link:https://tsinghua-gongjing.github.io/posts/bioinformatics_file_format.html
Previous:
Color for plot
Latest articles
Links
- ZhangLab , RISE database , THU life , THU info
- Data analysis: pandas , numpy , scipy
- ML/DL: sklearn , sklearn(中文) , pytorch
- Visualization: seaborn , matplotlib , gallery
- Github: me